- Visibility 37 Views
- Downloads 3 Downloads
- DOI 10.18231/j.ijmmtd.2024.068
-
CrossMark
- Citation
Prevalence, antimicrobial resistance patterns and ESBL resistance genotypes of Klebsiella pneumoniae Isolated from patients presenting with urinary tract infections at Mama Lucy Hospital, Kenya
Introduction
Klebsiella. pneumoniae is a gram-negative rod belonging to the Enterobacteriaceae family that causes infections in humans depending on its site.[1] This pathogen is known to cause pneumonia and urinary tract infections as well as other infections, including intra-abdominal infection, meningitis, pyogenic liver abscess, blood infection, and skin or soft tissue infections.[1] Nevertheless, this bacterium is a commensal of the digestive system and can also be found in the environment. This organism often causes infections, mostly among hospitalized and immunocompromised individuals treated with ß-lactam antibiotics. [2] Cephalosporin- and carbapenem-resistant Klebsiella pneumoniae strains have been reported in South Africa to cause several mortalities in Johannesburg. [3] The prevalence of ESBL genes in human hospital disease samples isolated from Klebsiellla pneumoniae in Ghana is 64%.[4] A study conducted in Cameroon reported that the percentage of ESBL-producing K. pneumoniae was 86.9%. [5] A study conducted in Kenya, ESBL-producing K. pneumoniae isolates were 92.2% and showed resistance to antibiotics, ceftriaxone (91.3%) amocillin/clavulanic acid (70.9%) cefepime (60.6%) nitrofurantoin (45.5%) azithromycin (22%) levofloxacin (12.6%) minocycline (11.8%) cefoxitin 9.4%.[6]
Urinary tract infections (UTIs) are the most common infections in primary care settings, affecting approximately 150 million people per year worldwide.[7] The World Health Organization has ranked resistance to antimicrobials among the 10 most serious global public health crises facing healthcare at this time. [8] The WHO flags K. pneumoniae as an important antimicrobial (RAM)-resistant bacteria because of its strong tendency to develop resistance to current antibiotics, such as penicillins, cephalosporins and quinolones. [9], [10], [11] Carbapenems are considered to be the most effective antibiotics for treating multidrug resistance (MDR); however, K. pneumoniae isolates can be resistant to all three or more drug classes. [12] A high morbidity of 404.6 million urinary tract infections was reported in 2019; for this reason, monitoring and research are needed to reduce this healthcare burden. [13] In Africa, the prevalence of ESBLs in Enterobacteriaceae has been researched at the local level in various countries, but there is no summarizing research on how prevalent ESBLs are on the continent, what types of genes are involved and where research is missing. [14]
Urinary tract infections pose a significant public health threat requiring significant economic implications. Due to the high empiric use of antibiotics for the treatment of UTI, antibacterial resistance of Enterobacteriaceae, specifically the main uropathogens Escherichia coli and K. pneumoniae, has significantly increased worldwide. [15] ESBL-producing Klebsiella strains are resistant to a number of antibiotics, such as aminoglycosides, fluoroquinolone, tetracyclines, chloramphenicol, trimethoprim + sulfamethoxazole, penicillins and cephalosporins.[16] Carbapenems, such as imipenem, meropenem, and ertapenem, can only be used to treat infections caused by ESBL strains,[17] although carbapenems should be avoided because they are used in cases of high antibiotic resistance infection.[18] A study conducted in low- and middle-income countries reported a dramatic increase in the resistance of K. pneumoniae to amoxcillin/ampicillin (80%) and co-trimoxazole (67%).[19]
In sub-Saharan Africa, the resistance of K. pneumoniae to third-generation cephalosporins has increased from 8 to 77%.[8] In Ethiopia, a study reported that K. pneumoniae isolated from urine samples presented 66.7% resistance to cotrimoxazole and 100% resistance to ampicillin.[20] In Gabon, a prevalence of 16.2% in patients infected by the urinary tract has been reported for K. pneumoniae, and the bacterium has shown significant resistance to beta-lactams, quinolones and cotrimoxazole.[21] In Nigeria, strong resistance of K. pneumoniae to ampicillin has been reported. [22] In Kenya, Kenyatta National Hospital (KNH) microbiology laboratory reported that isolated K. pneumoniae was resistant to all antibiotics used to treat childhood infections.[23] Over the past two decades, the prevalence of K. pneumoniae has been 23%, and a high resistance of more than 80% to penicillins, cephalosporins, macrolides, tetracyclines, sulfonamides, lincosamides and chloramphenicol has been reported in Kenya.[24] Affordable first-line agents such as ampicillin and gentamicin are unlikely to be clinically effective in a substantial proportion of infections. This has resulted in the increasing use of third-generation cephalosporins for the empirical treatment of serious infections.[19]
In Kenya, the multidrug resistance of K. pneumoniae has been reported to be greater than 80% for penicillins, chloramphenicol, cephalosporins, lincosamides, tetracyclines, macrolides and sulfonamides. Resistance to carbapenems was lowest at 23.2%, while resistance to amikacin, meropenem, aminoglycosides and quinolones was reported to be 21%, 7%, 49.2% and 41.3%, respectively. [24] This evidence shows that K. pneumoniae resistance is ever changing; thus, routine studies should be conducted to monitor it. There is also growing concern regarding the lack of new antibiotics, especially for multidrug-resistant gram-negative bacteria that produce ESBLs.[25]
This study therefore aims to help in understanding the trends of extended beta lactamase production in K. pneumoniae in UTI.
Materials and Methods
Study site
The study was carried out at Mama Lucy Kibaki Hospital between September 2023 and November 2023. Mama Lucy Hospital is a subcounty referral hospital providing comprehensive health care for both outpatients and inpatients within Nairobi City, Kenya. The hospital is located within Nairobi County between the Umoja-II and Komarock Estates of Nairobi, Kenya.
Study design
This was a cross-sectional study.
Study population
An estimated 400 participants aged between 18 years and 70 years were recruited into the study. The study targeted individuals presenting with UTI-like symptoms, including abdominal pain, frequent urination, back pain, cloudy urine, a burning sensation during urination, and a strong pungent smell. Those who consented to participate in the study were recruited while excluding those who did not consent and who were not able to produce urine or who were on antibiotic treatment.
Sample size determination
The sample size was determined using Fisher’s formula [26]
N=Z 2 pq/d 2
where n= Sample size, Z= 1.96 at the 95% confidence interval, where the prevalence of UTI of 31.6% was considered; Q=1-P, D=degree of accuracy 0.05 at 95%, thus n=3.84∗0.316∗0.6847/ (0.05)2, n=400
The sample size was calculated on the basis of the 31.6% prevalence of K. pneumoniae mentioned in previous study conducted in Ethiopia. [27]
Sampling criteria
Purposive sampling was used to select patients with UTIs.
Specimen collection
Sterile midstream urine samples were obtained from each consenting study participant under the instructions to urinate small amount into the toilet, then without stopping the flow of urine to med-stream urine in given sterile container up to 20mls. These samples were then sent to the CMR/KEMRI laboratory while kept 4oC. conditions for analysis within six hours of collection
Urinalysis
All 400 participants’ urine samples that were collected were subjected to macroscopy examination for color, smell, and consistency. Urine analysis was carried out using standard ComboStick 10 strips to assess the semiquantitative levels of leukocytes, pH, blood, nitrites, specific gravity and proteins in urine. [28]
Urine culture
Using a sterile calibrated plastic loop, approximately 10 µl urine samples were inoculated on both MacConkey agar and Cystine Lactose Electrolyte Deficient (CLED) media and incubated at 37 °C. for 18–24 hrs. The bacterial or fungal isolates were identified on the basis of their culture characteristics. The presumptive bacteria were confirmed using Gram stain and specific biochemical tests. [29]
Antimicrobial susceptibility test and ESBL detection
Approximately 0.5 McFarland’s standard pure bacterial isolates were inoculated to form a lawn on Mueller–Hinton agar. The antibiotic discs were distributed evenly on the agar surface and incubated at 37 °C. for 18–24 hrs. [30] The tested antimicrobials were ampicillin, cefotaxime, cefuroxime, ceftazidime, cefepime, imipenem, amoxicillin, clauvanalic acid, ceftriaxone, chloramphenicol, tetracycline, ciprofloxacin, sulfamethoxazole/trimethoprim and nitrofurantoin. The zones of inhibition were measured in millimeters by measuring the diameter of the zones using a ruler. The findings were compared to those obtained from controls followed guidelines for standard bacteria according to the Clinical and Laboratory Standards Institute. [31] The interpretation of the inhibition zone diameters was interpreted as susceptible, intermediate or resistant as per the Clinical and Laboratory Standards Institute guidelines. [31] The isolates that were resistant to cephalosporins and exhibited a synergy zone were further genetically analysed ESBL genes.
DNA extraction
The bacterial DNA extraction was performed by boiling method. The partial gene was amplified using specific primers ([Table 1]) in a 25 μL constituting of 5x buffer 5μl, template DNA 1 μl, DNTPs, MgCl2, taq polymerase, forward primer 0.5 μl, reverse primer 0.5 μl and nuclease-free water to 18μl under the following conditions; blaSHV at 940C for 30 seconds followed by 30 cycles at 94°C for 30 seconds, 50°C for 60 seconds, and 68°C for 1 min, with a final extension of 68°C for 5 min; blaTEM at 94°C for 30 seconds followed by 30 cycles of 94°C for 30 seconds, 50 °C for 60 seconds and 68°C for 1 min with a final extension at 68°C for 5 minutes, blaCTX-M at 94°C for 30 seconds followed by 30 cycles of 95°C for 30 seconds, 60°C for 60 seconds, 68°C for 1 min with a final extension at 68°C for 5 min and blaOXA at 94°C for 30 seconds followed by 30 cycles of 95°C for 30 seconds, 62°C for 60 seconds, 68° C for 1 min with a final extension at 68° C for 5 minutes. PCR amplification was confirmed via visualization with SYBR Safe DNA stain using gel electrophoresis. [32]
|
Gene |
Type |
Primer sequence |
Annealing temp |
Amplicon size (bp) |
Ref. |
|
SHV |
Fw |
5’-ACTATCGCCAGCAGGATC-3’ |
500 C |
356 |
|
|
|
Rev |
5’-ATCGTCCACCATCCACTG-3’ |
|
|
|
|
TEM |
Fw |
5’-GATCTCAACAGCGGTAAG-3’ |
500 C |
786 |
|
|
|
Rev |
5’-CAGTGAGGCACCTATCTC-3’ |
|
|
|
|
CTX- |
Fw |
5’-GTGATACCACTTCACCTC-3’ |
600 C |
255 |
|
|
M15 |
Rev |
5’-AGTAAGTGACCAGAATCAG-3’ |
|
|
|
|
OXA |
Fw |
5′-ATGAAAAACACAATACATATCAACTTCGC-3′ |
620 C |
820 |
|
|
|
Rev |
5′-GTGTGTTTAGAATGGTGATCGCATT-3′ |
|
|
|
Data and Statistical Analysis
Descriptive statistics was to analyse socio-demographic characteristics, epicollect excell sheet was used to record patients’ information, whonet was used to analyse antimicrobial susceptibility testing and excel was used for data recording and analysis
Results
Demographic characteristics of study participants
The participants aged from 18 years to 70 years were recruited in the study. The age group30-35(33.5%) was the most predominant followed by age groups 24-29 (25.5%), 36-41(15.5%),18-2 (10.25%), 42-47 (6.5%), 48-53 (3%), 54-59 (2%), 60-65 (1.75%), respectively and the least was the age range of ≥66 at (1.5%). Females had a high number of participants 309(77.25%) compared to males 91(22.75%), and married participants were dominant compared to unmarried participants with 311 (77.75%) and 89 (22.25%), respectively, self-employed were at 169(42.25%) followed by employed with 155(38.75%) and the last were unemployed with 76(19%). The participants with secondary school level were the most dominant with 197(49.25%) followed by tertiary level with 191(47.75%), primary level with 10(2.5%) and the last was non-educated with 2(0.5%) [Table 2].
|
Parameters |
Population n=400 |
Frequency % |
|
Age in years |
|
|
|
18-23 |
41 |
10.25% |
|
24-29 |
100 |
25% |
|
30-35 36-41 |
134 66 |
33.50% 16.50% |
|
42-47 |
26 |
6.50% |
|
48-53 |
12 |
3% |
|
54-59 |
8 |
2% |
|
60-65 |
7 |
1.75% |
|
66+ |
6 |
1.50% |
|
Sex |
|
|
|
Males |
91 |
22.75% |
|
Females |
309 |
77.25% |
|
Marital status |
|
|
|
Married |
311 |
77.75% |
|
Single |
89 |
22.25% |
|
Occupation |
|
|
|
Employed |
155 |
38.75% |
|
Self-employed |
169 |
42.25% |
|
Unemployed |
76 |
19% |
|
Education level |
|
|
|
Primary |
10 |
2.50% |
|
Secondary |
197 |
49.25% |
|
Tertiary |
191 |
47.75% |
|
non-educated |
2 |
0.50% |
Dipstick positive and negative urine culture
Among 400 participants positive growth culture was observed in 206(51.5%) while negative growth was observed in 194(48.5%) [Table 3]
|
Dipstick test |
Positive culture |
Negative culture |
Total |
|
Positive |
203(50.75%) |
193(48.25%) |
396 |
|
Negative |
3(0.75%) |
1(0.25%) |
4 |
|
Total |
206 |
194 |
400 |
Prevalence of Klebsiella pneumoniae among UTI patients
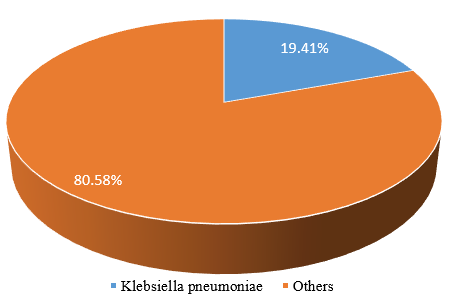
[Figure 1] shows the isolated K. pneumoniae and others. K. pneumoniae isolates were 40(19.41%) while other isolates were 166(80.58%) from participants enrolled in this study.
Antimicrobial susceptibility patterns
From a total of 40 K. pneumoniae isolate that were tested, high levels of resistance was observed in Ampicillin (AMP) with 84.37%, followed by Ceftriaxone (CTR) and Cefotaxime (CTX) that had equal proportion of resistance 40.62%, Sulfamethoxazole-trimethoprim (COT) with 37.50%, Cefuroxime (CXM) with 34.37%, Tetracycline (TE) with 31.25%, Amoxicilin-clavulanic (AMC) with 28.12%, Ciprofloxacin (CIP) with 21.90%, Cefepime (CPM) with 12.50%, Nitrofurantoin (NIT) with 6.25% and the low resistance was observed in Chloramphenicol(C) with 3.12%. None of the isolates showed resistance to Imipenem (IPM). K. pneumoniae isolates were sensitive to Chloramphenicol 96.87%, Imipenem 93.75%, and Nitrofurantoin 90.62%, Ceftriaxone, Cefotaxime, Cefepime and Sulfamethoxazole-trimethoprim showed an equal sensitivity of 53.12%, Ciprofloxacin and Cefuroxime with 50%, and Tetracycline 46.87%. None of the isolate showed sensitivity to Amoxicilin-clavulanic. However multi-drug resistance (MDR) was observed to Cefotaxime, Ampicilin, Chloramphenicol, Imipenem and Cefuroxime (31.25%).[Table 4]
|
|
Antimicrobial Susceptibility patterns |
|
|
|
Antibiotics |
R (%) |
I (%) |
S (%) |
|
Ampicillin |
84.37% |
3.12% |
12.50% |
|
Amoxicilin-clavulanic |
28.12% |
71.90% |
0% |
|
Cefuroxime |
34.37% |
15.62% |
50% |
|
Ceftriaxone |
40.62% |
6.25% |
53.12% |
|
Cefotaxime |
40.62% |
6.25% |
53.12% |
|
Cefepime |
12.50% |
31.25% |
53.12% |
|
Imipenem |
0% |
6.32% |
93.75% |
|
Ciprofloxacin |
21.90% |
28.12% |
50% |
|
Sulfamethoxazole-trimethoprim |
37.50% |
9.37% |
53.12% |
|
Chloramphenicol |
3.12% |
0% |
96.87% |
|
Tetracycline |
31.25% |
21.87% |
46.87% |
|
Nitrofurantoin |
6.25% |
3.12% |
90.62% |
Among the 40 K. pneumoniae isolates, 30 (75%) were ESBL-positive. [Figure 2]

Resistance genes of Klebsiella pneumoniae
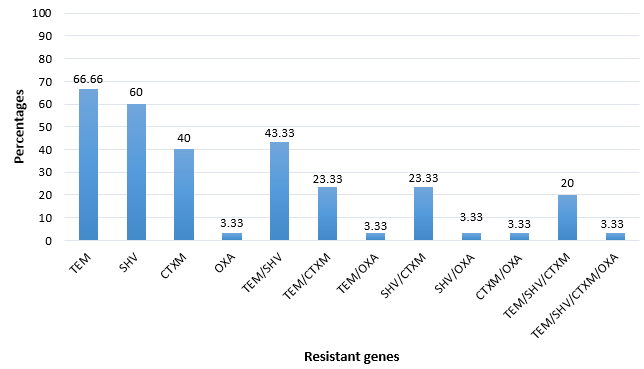
From the susceptibility profiles, those bacterial isolates that were β-lactamase resistant, their associated genes were determined. The frequency of these genes was; blaTEM (66.7%) followed blaSHV (60%), blaCTX-M (40%) with blaOXA (3.33%) as the least β-lactamase genes. The co-existence of multiple bla genes in a bacterial isolate was observed with blaTEM/SHV (43.33%) combined being the most common with blaTEM/SHV/CTXM/OXA as the least drug-resistant genes (3.33%). [Figure 3]
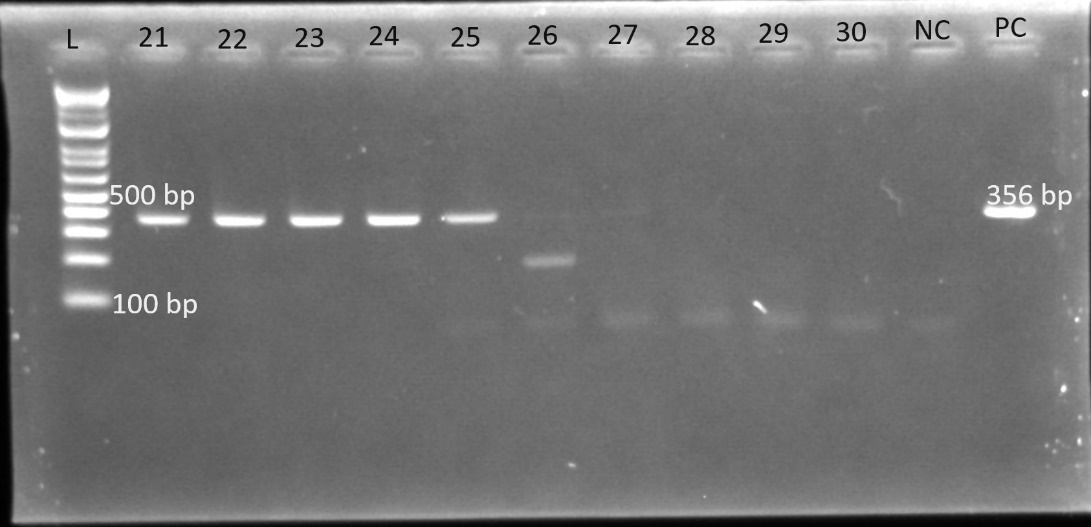
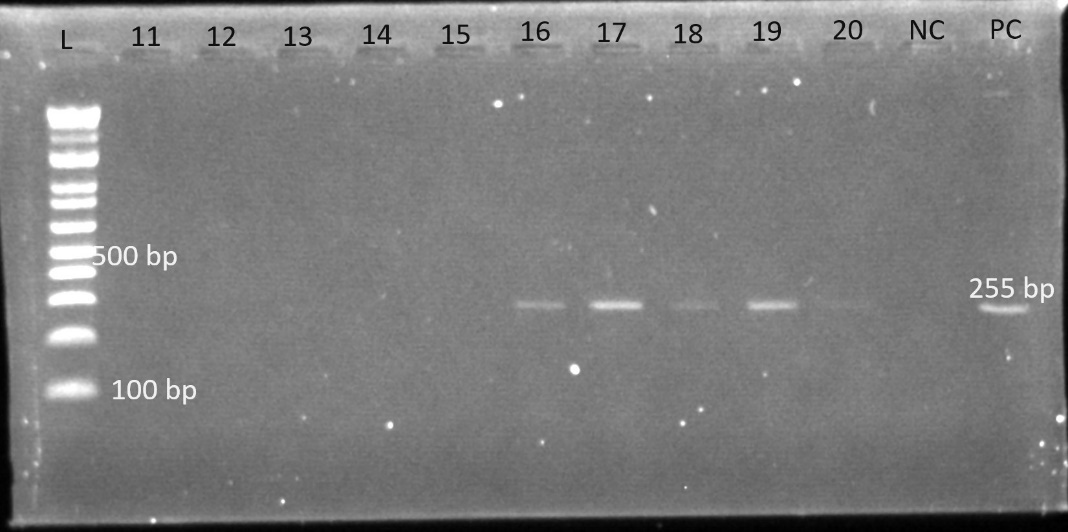
Gel electrophoresis was used to identify the bands. The gel plates below are representative electrophoresis gel images.
Discussion
Socio-demographic characteristics of the participants
In this study the majority of participants were females compared to males and the predominant age range was 30-35years ([Table 1]). In Kenya reported the same findings, where the number of female participants was greater than that of male participants.[34] Another study conducted in Nigeria, reported the same findings, where the majority of participants were females (66%) rather than males (34%). [35] The findings of a study carried out in Uganda on UTIs revealed that there were more female participants (37.5%) than male participants [36], [21] reported that most participants were females (61.7%) and males (38.3%). Among women and men, urinary tract infection affects all age groups, with a higher incidence occurring among sexually active women. [37] Having external anatomical structure of reproductive organs that may favour the infection and shorter urethra closer to anus, expose females to UTI than males, because the bacteria ascendance from anus can easily enter through the urethra and invade the bladder. [37] Sexually intercourse is more common at 30 s, and at high frequency, this may be the cause. A contradictory study conducted by. [35] revealed that the majority of participants were in the age range of 22–26 years (36%). Other studies contradicted the findings of,[36], [38], [39] who reported that the most prevalent age ranges were 16–30 years (37.9%), 20–29 years (32.6%), and >45 years (25%), respectively. In Ethiopia, a study on the prevalence of bacterial urinary tract infections reported a high occurrence of the disease among those in the 20–69, years groups,[27] which is in agreement with the findings of the current study, where people in the mentioned age interval were more prone to UTIs. Compared with single individuals, married individuals are reported to be more affected by UTIs,[40] which may be due to the frequency of transmission of infection among couples, which contributes to a significant number of married women having UTIs than men do.[27]
Prevalence of Klebsiella pneumoniae among UTI patients
The prevalence of Klebsiella pneumoniae reported in the present study was 19.41% ([Figure 1] ), which is lower than the prevalence reported in a previous study carried out in Kenya which reported prevalence of 33.3% Klebsiella pneumonia.[34] In Kenya study conducted reported that 29% was contributed to Klebsiella pneumoniae in UTI.[41] Another study carried out in Bangladesh reported a prevalence of 21.6% Klebsiella pneumonia.[42] A study conducted in Kenya showed a prevalence of Klebsiella pneumoniae which stood at 8.9%.[43] In Uganda, a prevalence of 11.6% was reported.[36] Among the Klebsiella species contributing to UTIs in Northwest Ethiopia, Klebsiella pneumoniae was the most common isolate, with more than twice the number of other Klebsiella species isolated. [38] its prevalence was 65.8%, which is higher than what the current study has determined. Similarly, a high prevalence of Klebsiella pneumoniae was observed in the U.S. and represented more than half of all Klebsiella spp. isolated from urine samples. [44] This may be due to poor hygiene and wiping back to front rather than front to back. [45]
Among community uropathogen isolates in Gabon, Klebsiella pneumoniae accounts for approximately 16.2% of the total prevalence of UTIs, making it the second most common isolated microorganism among patients.[21] Another study conducted in Saudi Arabia reported that Klebsiella pneumoniae was the most commonly observed isolate among its genera, accounting for 97.4% of the Klebsiella spp. identified in urine.[46] With its ability to invade soft tissues, Klebsiella pneumoniae uses its type 1 fimbriae in the attachment and invasion of bladder epithelial cells, which may also favour the formation of biofilms supporting its long survival in the bladder, leading to its high prevalence in chronic UTIs.[47] Among the Klebsiella pneumoniae isolates, the high prevalence of ESBL-producing Klebsiella pneumoniae was 75% (Figure.2).
A study conducted in Cameroon reported a prevalence of Klebsiella pneumoniae of 82.4%.[5] In Turkey, a similar study on urinary tract infection reported a high prevalence of ESBL-producing Klebsiella pneumoniae, which was 78.8%.[48] Another study carried out in Nigeria reported a prevalence of 31% for ESBL-producing Klebsiella pneumonia,[35] which is very low compared with the prevalence reported in the current study. In the Tertiary Care Hospital of Central Nepal, the total reported prevalence of K. pneumoniae was 223 (11.70%), 19% of which were ESBL producers.[49] In Iran, the percentage of ESBL-harboring K. pneumoniae was 13.5%,[50] while it was reported to be 31% in a study carried out in Nigeria. [35] In Kenya, a study carried out in two referral hospitals reported a high prevalence of ESBL-producing K. pneumoniae, which was 92.8%.[6] K. pneumoniae was reported as one of the top ESBL producers among Enterobacteriaceae, which supports its high occurrence in the chronic phase of urinary tract infection.[51]
Antimicrobial susceptibility patterns and ESBL producing K. pneumoniae
In the 20th century, the discovery of antibiotics has played a significant role in the treatment of microbial diseases [52], and antimicrobial resistance resulting from the high selection of antibiotics for treatment and their misuse and overuse has presented a global public health challenge for health systems in recent decades. [53] The present study revealed high resistance (84.37%) of this uropathogen to ampicillin and MDR phenotypes to cefotaxime, ampicillin, chloramphenicol, and cefuroxime, whereas the highest sensitivity was observed for chloramphenicol, imipenem, and nitrofurantoin ([Table 2]).
Similar findings were reported in India where K. pneumoniae showed the highest resistance to ampicillin (75.6%), nitrofurantoin (73.1%), and cefuroxime.[54] Additionally, all ESBL-producing K. pneumoniae strains were reported to be MDR, while they showed high resistance to nitrofurantoin and contrimonazole.[54] In Morocco, differences in resistance for both non-ESL-producing K. pneumoniae and ESBL-producing K. pneumoniae were detected for sulfamethoxazole-trimethoprim (61%, 89%), ciprofloxacin (32%, 84%), gentamicin (21%, 89%), and amikacin (11%, 50%), with the highest resistance reported for ESBL-producing K. pneumoniae rather than non-ESL-producing K. pneumonia. [55] The Klebsiella pneumoniae ESBL producer was sensitive to imipenem (96%), as reported by [5] in Cameroon. In the Kurdistan Region of Iraq, K. pneumoniae has been reported to be highly resistant to ampicillin (96.9%), ceftriaxone (65.8%), and cefepime (60.8%), whereas the highest sensitivity was observed for ertapenem (93.8%), and 82.3% was attributed to mipenem. [56]
In Egypt, a study conducted in a tertiary care hospital showed that 90% of ESBL-producing Klebsiella pneumoniae strains were resistant to Sulmethoxazole/trimethoprim, 70% were resistant to amoxicillin/clavulanate, 63.3% were resistant to cefotaxime, 40% were resistant to cefepime, 46.7% were resistant to ceftriaxone, and the least resistance is observed for imipenem.[57] Another study conducted in Italy reported the broad resistance of K. pneumoniae to penicillins and cephalosporins, whereas high resistance of ESBL-producing K. pneumoniae to carbapenem antibiotics was reported.[58] Similar findings revealed 100% resistance to ampicillin, whereas 70–80% resistance was reported for first-, second-, and third-generation cephalosporins. However, high sensitivity to imipenem has been reported. [59]
The present study reported the high resistance to ampicillin, in agreement with study conducted in Bangladesh, the majority of K. pneumoniae strains (82%) are MDR and were resistant to antibiotics, including β-lactam antibiotics, aminoglycosides, carbapenems, and ciprofloxacin [60] The uniform sensitivity of K. pneumoniae to imipenem was reported in a study carried out in India, but the study also revealed high susceptibility to β-lactamase combined drugs (67–81%) and aminoglycosides (62–76%); however, the lowest sensitivity was observed for third-generation cephalosporins (14–24%) and non-β-lactam antibiotics, including nitrofurantoin (57%), fluoroquinolones (29–57%), piperacillin (19–23%), and aztreonam. [61]
Another study conducted in Iran revealed that all ESβL-producing K. pneumoniae strains were sensitive to imipenem and meropenem, and the isolates were resistant to aztreonam. Significantly high resistance to amoxicillin (100%), cefotaxime (50%), and gentamicin (42.3%) was observed, whereas the least resistance to imipenem (15.9%) and meropenem (11.8%) was observed. [62] Approximately 99% of ESβL-producing K. pneumoniae were resistant to sulfonamides; 81% were resistant to quinolones, whereas 79% were resistant to aminoglycosides. [51] In the last two decades, a significant decrease in the susceptibility of K. pneumoniae to third-generation cephalosporins and ciprofloxacin was reported, with sensitivities of 83.6% and 81.6% attributed to cefotaxime and ciprofloxacin, respectively, in 2012. [63]
Another study conducted in Kenya reported that, compared with other isolates, Klebsiella pneumoniae was more resistant to nitrofurantoin. [64] A study conducted in Benin reported that ESBL-producing Klebsiella pneumoniae was highly resistant to amoxicillin (79.07%), cefotaxime (53.48%), and trimethoprim/sulfamethoxazole combinations (86.05%). [65] In Bagdad, Iraq, high resistance of K. pneumoniae was detected to antibiotics such as ampicillin (100%), cefixime (73.8%), cefuroxime (71.05%), and ceftazidime (65.79%), and intermediate resistance was reported to antibiotics such as nitrofurantoin, sulfamethoxazole-trimethoprim, and ceftriaxone, while the lowest resistance was observed to antibiotics, including imipenem, meropenem, and ciprofloxacin. [66] In Ethiopia, K. pneumoniae has been isolated from other Enterobacteriacieae and has shown high resistance to cotrimoxazole (91.7%) and chloramphenicol (66.7%), whereas other types of resistance have been reported for ciprofloxacin (45.8%), norfloxacin (45.8%), and 25% resistance to gentamicin. In terms of MDR, this uropathogen (K. pneumoniae) has 58% MDR among other Enterobacteriacieae. [67]
Other findings reported in Gaza in Palestine, the high resistance of K. pneumoniae was observed to antibiotics such as Cefotaxime, ampicillin, and Sulfamethoxazole-trimethoprim.[68] similarly, another study reported the high resistance of K. pneumoniae to ampicillin, Sulfamethoxazole-trimethoprim, Cefotaxime, piperacillin, levofloxacin, gentamicin, ceftazidime, ceepime, and aztreonam, although, the moderate resistance was observed to ciprofloxacin and ceftriaxone, while the least resistance was detected for imipenem. [69] It was also observed in another study that K. pneumoniae isolated from was 90% MDR and all MDR K. pneumoniae were ESBL- producers. [70]
Extended-spectrum β-lactamase resistance genes in Klebsiella pneumoniae
One of challenges in the use of β-lactam antibiotics for treatment of bacterial infections is the production of Extended-spectrum β-lactamases enzyme by majority pathogens of which K. pneumoniae was ranked among the top. However, the production of this enzyme (ESBL) was found to be a result of gene expression from bacterial genome. [71] In this study, we have observed a high occurrence of blaTEM and blaSHV, the moderate occurrence of blaCTX-M, and the least occurrence was observed to blaOXA. The combination of bla genes was also studied ([Figure 1]). [72] has reported the similar findings where blaTEM (49.4%) was reported as the most common genotype detected, in contrast, blaSHV was observed as the least detected genotype in K. pneumoniae.
The same bla genes reported in a study carried out in Malaysia contradicted the findings of the present study: blaSHV was detected in 46 K. pneumoniae isolates for blaSHV, 19 isolates for blaCTX-M, 5 isolates for blaOXA, and 4 isolates for blaTEM. [73] In Southwest Nigeria, different percentages of K. pneumoniae bla genes have been reported, and all the genes detected in the present study were also detected in Nigeria at different percentages. The bla genes detected included blaTEM (47.7%), blaCTX-M (43.8%), blaSHV (39.8), and blaOXA, which were the least frequently detected and represented 27.3% of all genes. [74] Another study revealed that blaCTX-M occurred in 30% of isolates, whereas other bla genes were not detected. [75]
Similar bla genes were reported in a study conducted in Egypt 10% of Klebsiella pneumoniae isolates was for blaSHV and 53.3% for blaCTX-M. [57] Klebsiella pneumoniae isolated from hospital in Benin reported the occurrence of blaSHV and blaCTX-M. [65] Differently, a study carried out in China, showed the high occurrence of blaSHV which was the most prevalent, and the second prevalent was blaTEM, while the least prevalent reported bla gene was blaCTX-M with zero to blaOXA. [76] Again, another study in China, reported the high prevalence of blaTEM in ESBL-KP which stood at 69.3%, blaCTX-M was 45.5% and the least observed was blaSHV which stood at 4.5%, no blaOXA was also reported for this study.[77] In Iran, ESBL-producing K. pneumoniae showed the presence of 59.3% for blaTEM while the presence of the combination of blaCTX-M/ blaTEM was observed at 33.3%.[78] In Nepal, the overall prevalence of blaCTX-M was 89.62% in which K. pneumoniae had 78.94%. [79] In hypervirulent ESBL-producing K. pneumoniae, blaSHV (63.8), while 59% and 58.1% were attributed to blaTEM and blaCTX-M respectively. [80]
ESBL bla genes were also found and described in 32 ESBL-producing K. pneumoniae strains, where blaCTX-M genes were detected in 20 isolates. blaSHV genes were detected in 2 isolates, whereas combinations of blaCTX-M genes and blaSHV genes were detected in 9 isolates, with an unknown gene that was observed in 1 isolate. [81] The overall prevalence rates reported for blaTEM, blaCTX-M, and blaSHV were 86%, 78%, and 28%, respectively, of which ESBL-producing K. pneumoniae carried 34% for blaTEM, 31% for blaCTX-M, and 26.1% for blaSHV genes. [82] In Kenya, the predominant ESBL genes reported for K. pneumoniae were blaTEM, which stood at 89%; blaSHV, which stood at 82.7%; blaOXA, which stood at 76.4%; and blaCTX-M, which stood at 72.5%. In addition, the presence of the blaSHV, blaOXA, and blaTEM genes were associated with MDR. [6]
Another contradictory study was carried out in Kenya, where a high occurrence of blaCTX-M genes was observed, followed by blaTEM, and the least detected bla gene was blaOXA.[41] A study in which samples from both Kenya and Uganda were collected detected K. pneumoniae resistance bla genes, which included blaCTX-M genes, blaTEM genes, and blaOXA, the K. pneumoniae strains with these genes were 100% resistant to 3 more antibiotics, revealing the trend of MDR for these bacteria and predicting future pandemics resulting from this threat if nothing is done.[83] In Nigeria, the two K. pneumoniae isolates harboured both blaSHV and blaCTX-M of the blaCTX-M-1 group, the third K. pneumoniae harboured only blaCTX-M of the blaCTX-M-1 group, and the study revealed the critical threat of the increase in carbapenem-resistant K. pneumoniae resulting from coharbouring both blaCTX-M of the blaCTX-M-1 group and blaSHV genes. [84]
In Eastern province, South Africa, ESBL-producing K. pneumoniae (n=139) harboured the high prevalence of blaSHV (n=22) for the single ESBL genes, but also showed blaTEM (n=5), while blaCTX-M was observed at low frequency in this category, for two or more ESBL genes, K. pneumoniae had 78/139 for blaTEM + blaSHV + blaCTX-M, 12/139 for blaTEM + blaSHV, 6/139 for blaCTX-M + blaSHV, and 4/139 for blaCTX-M + blaTEM, no blaOXA gene was observed in the study.[85] A study carried out in three countries (Egypt, Saudi Arabia, and Sudan) revealed that blaCTX-M was detected in all K. pneumoniae isolates while blaTEM was reported at 66.7%, the presence of these genes highlighted the high MDR in mentioned MDR to the last line antibiotics.[86] Another similar study reported the high prevalence of blaTEM genes which stood at 57.1%, blaCTX-M had 28.6%, some genes were occurred in combination within K. pneumoniae genotype, and include blaSHV and blaCTX-M which stood at 14.3%, blaTEM, blaSHV, and blaCTX-M stood at 14.3%.[87]
Limitations
Due to the lack of resources the advanced molecular method which show whether the genes found were plasmid or chromosomal associated was not done.
Conclusion
The prevalence of K. pneumoniae was 19.40%, 75% of which were ESBL producers. This uropathogen showed high resistance to ampicillin and high susceptibility to chloramphenicol, imipenem, and nitrofurantoin. Multidrug resistance (MDR) to antibiotics such as cefotaxime, ampicillin, chloramphenicol, and cefuroxime has been detected. The observed ESβL resistance genes in K. pneumoniae included blaTEM, blaSHV, blaCTX-M, and blaOXA, with blaTEM being the most prevalent. The successful completion of this study highlighted the emerging resistance of K. pneumoniae among urinary tract-infected patients. Thus, this study recommends the need for that healthcare facilities for routine laboratory testing for ESBL phenotypic and molecular UTI diagnoses to guide clinical treatment of UTI patients and AMR need for regular surveillance for the emergence and spread of ESBL among UTI-causing KP.
Ethical Consideration
Ethical clearance was granted by Jomo Kenyatta University of Agriculture and Technology ethical committee JKU/02316/0821. The study also obtained approval from the National Commission for Science, Technology & Innovation (Nacosti) Ref No:237148.
Conflicts of Interest
The authors declare no conflicts of interest.
Source of Funding
None.
Acknowledgments
The authors would like to acknowledge KEMRI-CMR Laboratory staff for their cooperation during the study period as well as the study participants for their participation in the study.
References
- M Ostojic, M Hubana, M Cvetnić, M Benić, Z Cvetnić. Antimicrobial resistance of Klebsiella pneumoniae strains isolated from urine in hospital patients and outpatients. Arch Biotechnol Biomed 2021. [Google Scholar] [Crossref]
- RM Martin, MA Bachman. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018. [Google Scholar] [Crossref]
- NM Mbelle, C Feldman, JO Sekyere, NE Maningi, L Modipane, SY Essack. Pathogenomics and Evolutionary Epidemiology of Multi-Drug Resistant Clinical Klebsiella pneumoniae Isolated from Pretoria, South Africa. Sci Rep 2020. [Google Scholar] [Crossref]
- JK Calland, K Haukka, SW Kpordze, A Brusah, M Corbella, C Merla. Population structure and antimicrobial resistance among Klebsiella isolates sampled from human, animal, and environmental sources in Ghana: a cross-sectional genomic One Health study. Lancet Microbe 2023. [Google Scholar]
- T Jean, CO Ebongue, C Yadufashije, L Kojom, D Adiogo. Phenotypic Characteristics of Klebsiella pneumoniae Extended Spectrum β-Lactamases Producers Isolated in Hospitals in the Littoral Region, Cameroon. Eur. Eur J Clin Microbiol 2020. [Google Scholar]
- SM Maveke, GO Aboge, LW Kanja, AO Mainga, N Gachau, BW Muchira. Phenotypic and Genotypic Characterization of Extended Spectrum Beta-Lactamase-Producing Clinical Isolates of Escherichia coli and Klebsiella pneumoniae in Two Kenyan Facilities: A National Referral and a Level Five Hospital. Int J Microbiol 2024. [Google Scholar] [Crossref]
- C Yadufashije, L Muhimpundu, E Munyeshyaka, J Mucumbitsi. The human vaginal microbial community dysbiosis contributes to the urinary tract infections during pregnancy: Case study of Gisenyi District Hospital. Rwanda Asian J Med Sci 2021. [Google Scholar]
- . WHO. National Action Plan on Prevention and Containment of Antimicrobial Resistance, 2017–2022. Regional Office for Africa: Brazzaville, Republic of Congo. . [Google Scholar]
- L Chen, B Mathema, KD Chavda, FR Deleo, RA Bonomo, BN Kreiswirth. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 2014. [Google Scholar]
- SP Henson, CJ Boinett, MJ Ellington, N Kagia, S Mwarumba, S Nyongesa. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol 2017. [Google Scholar]
- D Rawat, D Nair. Extended-spectrum β-lactamases in Gram Negative Bacteria. J Glob Infect Dis 2010. [Google Scholar]
- AP Magiorakos, A Srinivasan, RB Carey. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012. [Google Scholar]
- Z Zeng, J Zhan, K Zhang, H Chen, S Cheng. Global, regional, and national burden of urinary tract infections from 1990 to 2019: an analysis of the global burden of disease study 2019. World J Urol 2022. [Google Scholar]
- V Storberg. ESBL-producing Enterobacteriaceae in Africa - a non-systematic literature review of research. Infect Ecol Epidemiol 2008. [Google Scholar] [Crossref]
- A Mazzariol, A Bazaj, G Cornaglia. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. J Chemother 2017. [Google Scholar]
- ME Falagas, DE Karageorgopoulos. Extended-spectrum beta-lactamase-producing organisms. J Hosp Infect 2009. [Google Scholar]
- S Nathisuwan, DS Burgess, JS Lewis. Extended-spectrum beta-lactamases: epidemiology, detection, and treatment. Pharmacotherapy 2001. [Google Scholar]
- KK Kumarasamy, MA Toleman, TR Walsh, J Bagaria, F Butt, R Balakrishnan. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 2010. [Google Scholar]
- IJ Stanley, H Kajumbula, J Bazira, C Kansiime, IB Rwego, BB Asiimwe. Multidrug resistance among Escherichia coli and Klebsiella pneumoniae carried in the gut of out-patients from pastoralist communities of Kasese district, Uganda. PLoS One 2018. [Google Scholar] [Crossref]
- A Bitew, T Molalign, M Chanie. Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC Infect Dis 2017. [Google Scholar] [Crossref]
- YM Ndzime, R Onanga, RFK Kassa, M Bignoumba, PPM Nguema, A Gafou. Epidemiology of Community Origin Escherichia coli and Klebsiella pneumoniae Uropathogenic Strains Resistant to Antibiotics in Franceville, Gabon. Infect Drug Resist 2021. [Google Scholar] [Crossref]
- S Pokharel, S Raut, B Adhikari. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob Health 2019. [Google Scholar] [Crossref]
- . Ministry of Health Kenya issues new guidelines targeting multidrug resistant pneumonia. 2022. [Google Scholar]
- OE Apondi, OC Oduor, BK Gye, MK Kipkoech. High prevalence of multi-drug resistant klebsiella pneumoniae in a tertiary teaching hospital in Western Kenya. Afr J Infect Dis 2016. [Google Scholar]
- DO Ogbolu, OAT Alli, MA Webber, AS Oluremi, OM Oloyede. CTX-M-15 is Established in Most Multidrug-Resistant Uropathogenic Enterobacteriaceae and Pseudomonaceae from Hospitals in Nigeria. Eur J Microbiol Immunol (Bp) 2018. [Google Scholar]
- J Charan, T Biswas. How to calculate sample size for different study designs in medical research?. Indian J Psychol Med 2013. [Google Scholar]
- Y Gebretensaie, A Atnafu, S Girma, Y Alemu, K Desta. Prevalence of Bacterial Urinary Tract Infection, Associated Risk Factors, and Antimicrobial Resistance Pattern in Addis Ababa, Ethiopia: A Cross-Sectional Study. Infect Drug Resist 2023. [Google Scholar] [Crossref]
- V Kavuru, T Vu, L Karageorge, D Choudhury, R Senger, J Robertson. Dipstick analysis of urine chemistry: benefits and limitations of dry chemistry-based assays. Postgrad Med 2020. [Google Scholar]
- P Pradhan, J P Tamang. Phenotypic and Genotypic Identification of Bacteria Isolated From Traditionally Prepared Dry Starters of the Eastern Himalayas. Front Microbiol 2019. [Google Scholar] [Crossref]
- M Worku, G Belay, A Tigabu. Bacterial profile and antimicrobial susceptibility patterns in cancer patients. PLoS One 2022. [Google Scholar] [Crossref]
- . Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100-S19. . [Google Scholar]
- EN Cunha, MFB Souza, DCF Lanza, JPMS Lima. A low-cost smart system for electrophoresis-based nucleic acids detection at the visible spectrum. PLoS One 2020. [Google Scholar] [Crossref]
- MM Gharrah, AM El-Mahdy, RF Barwa. Association between Virulence Factors and Extended Spectrum Beta-Lactamase Producing Klebsiella pneumoniae Compared to Nonproducing Isolates. Interdiscip Perspect Infect Dis 2018. [Google Scholar] [Crossref]
- JW Maina, FG Onyambu, PS Kibet, AM Musyoki. Multidrug-resistant Gram-negative bacterial infections and associated factors in a Kenyan intensive care unit: a cross-sectional study. Ann Clin Microbiol Antimicrob 2023. [Google Scholar] [Crossref]
- KC Mofolorunsho, HO Ocheni, RF Aminu, CA Omatola, OO Olowonibi. Prevalence and antimicrobial susceptibility of extended-spectrum beta lactamases-producing Escherichia coli and Klebsiella pneumoniae isolated in selected hospitals of Anyigba, Nigeria. Afr Health Sci 2021. [Google Scholar]
- M Odoki, AA Aliero, J Tibyangye, JN Maniga, E Wampande, CD Kato. Prevalence of Bacterial Urinary Tract Infections and Associated Factors among Patients Attending Hospitals in Bushenyi District, Uganda. Int J Microbiol 2019. [Google Scholar] [Crossref]
- TA Rowe, M Juthani-Mehta. Urinary tract infection in older adults. Aging Health 2013. [Google Scholar] [Crossref]
- A Ameshe, T Engda, M Gizachew. Antimicrobial Resistance Patterns, Extended-Spectrum Beta-Lactamase Production, and Associated Risk Factors of Klebsiella Species among UTI-Suspected Patients at Bahir Dar City, Northwest Ethiopia. Int J Microbiol 2022. [Google Scholar] [Crossref]
- WD Seifu, AD Gebissa. Prevalence and antibiotic susceptibility of Uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital. BMC Infect Dis 2018. [Google Scholar] [Crossref]
- A Al-Badr, G Al-Shaikh. Recurrent Urinary Tract Infections Management in Women: A review. Sultan Qaboos Univ Med J 2013. [Google Scholar]
- A Muraya, C Kiyaga, S Kiyaga, HJ Smith, C Kibet, MJ Martin. Antimicrobial Resistance and Virulence Characteristics of Klebsiella pneumoniae Isolates in Kenya by Whole-Genome Sequencing. Pathogens 2022. [Google Scholar] [Crossref]
- S Chakraborty, K Mohsina, PK Sarker, MZ Alam, MI Abdul Karim, SM Abu Sayem. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Period Biol 2016. [Google Scholar]
- S Kiiru, J Maina, J Katana, J Mwaniki, BB Asiimwe, SE Mshana. Bacterial etiology of urinary tract infections in patients treated at Kenyan health facilities and their resistance towards commonly used antibiotics. PLoS One 2023. [Google Scholar] [Crossref]
- KS Kaye, V Gupta, A Mulgirigama, AV Joshi, G Ye, NE Scangarella-Oman. Prevalence, regional distribution, and trends of antimicrobial resistance among female outpatients with urine Klebsiella spp. isolates: a multicenter evaluation in the United States between. Antimicrob Resist Infect Control 2011. [Google Scholar] [Crossref]
- S Persad, S Watermeyer, A Griffiths, B Cherian, J Evans. Association between urinary tract infection and postmicturition wiping habit. Acta Obstet Gynecol Scand 2006. [Google Scholar]
- YA Almutawif, HMA Eid. Prevalence and antimicrobial susceptibility pattern of bacterial uropathogens among adult patients in Madinah, Saudi Arabia. BMC Infect Dis 2023. [Google Scholar] [Crossref]
- MES Guerra, G Destro, B Vieira, AS Lima, LFC Ferraz, AP Hakansson. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Front Cell Infect Microbiol 2022. [Google Scholar] [Crossref]
- S Alkan, II Balkan, S Surme, OF Bayramlar, SY Kaya, R Karaali. Urinary tract infections in older adults: associated factors for extended-spectrum beta-lactamase production. Front Microbiol 2024. [Google Scholar] [Crossref]
- SN Mahaseth, RK Sanjana, BK Jha, K Pokharel. Prevalence of Extended Spectrum Beta-Lactamase Producing Escherichia coli and Klebsiella pneumoniae Isolated from Urinary Tract Infected Patients Attending Tertiary Care Hospital of Central Nepal. J Coll Med Sci Nepal 2019. [Google Scholar]
- SB Zadeh, P Shakib, MR Zolfaghari, AF Sheikh. Prevalence of Escherichia coli and Klebsiella pneumoniae, Producing Extended-Spectrum Beta-Lactamase (ESBLs) from Clinical Specimen in Khuzestan, Iran. Gene Cell Tissue 2021. [Google Scholar] [Crossref]
- E Müller-Schulte, MN Tuo, C Akoua-Koffi, F Schaumburg, SL Becker. High prevalence of ESBL-producing Klebsiella pneumoniae in clinical samples from central Côte d'Ivoire. Int J Infect Dis 2020. [Google Scholar] [Crossref]
- WA Adedeji. The treasure called antibiotics. Ann Ib Postgrad Med 2016. [Google Scholar]
- MA Salam, MY Al-Amin, MT Salam, JS Pawar, N Akhter, AA Rabaan. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare (Basel) 2023. [Google Scholar] [Crossref]
- MG Manjula, GC Math, K Nagshetty, SA Patil, SM Gaddad, CT Shivannavar. Antibiotic Susceptibility Pattern of ESβL Producing Klebsiella pneumoniae Isolated from Urine Samples of Pregnant Women in Karnataka. J Clin Diagn Res 2014. [Google Scholar]
- M C El Bouamri, L Arsalane, Y El Kamouni, S Zouhair. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: A retrospective study from a university hospital in Morocco, North Africa. Afr J Urol 2015. [Google Scholar]
- IA Naqid, NR Hussein, AA Balatay, KA Saeed, HA Ahmed. The Antimicrobial Resistance Pattern of Klebsiella pneumoniae Isolated from the Clinical Specimens in Duhok City in Kurdistan Region of Iraq. J Kermanshah Univ Med Sci 2020. [Google Scholar] [Crossref]
- OI Ahmed, SA El-Hady, TM Ahmed, IZ Ahmed. Detection of bla SHV and blaCTX-M genes in ESBL-producing Klebsiella pneumoniae isolated from Egyptian patients with suspected nosocomial infections. J Med Genet 2013. [Google Scholar]
- B Santella, M Boccella, V Folliero, D Iervolino, P Pagliano, L Fortino. Antimicrobial Susceptibility Profiles of Klebsiella pneumoniae Strains Collected from Clinical Samples in a Hospital in Southern Italy. Can J Infect Dis Med Microbiol 2024. [Google Scholar] [Crossref]
- A Varghese, S George, R Gopalakrishnan, A Mathew. Antibiotic Susceptibility Pattern of Klebsiella pneumoniae Isolated from Cases of Urinary Tract Infection in a Tertiary Care Setup. J Evol Med Dent Sci 2016. [Google Scholar]
- P Aminul, S Anwar, MA Molla, MRA Miah. Evaluation of antibiotic resistance patterns in clinical isolates of Klebsiella pneumoniae in Bangladesh. Biosafety Health 2021. [Google Scholar]
- AK Singh, S Jain, D Kumar, RP Singh, H Bhatt. Antimicrobial susceptibility pattern of extended-spectrum beta- lactamase producing Klebsiella pneumoniae clinical isolates in an Indian tertiary hospital. J Res Pharm Pract 2015. [Google Scholar]
- D Mansury, M Motamedifar, J Sarvari, B Shirazi, A Khaledi. Antibiotic susceptibility pattern and identification of extended spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae from Shiraz, Iran. Iran J Microbiol 2016. [Google Scholar]
- WP Lin, JT Wang, SC Chang, FY Chang, CP Fung, YC Chuang. The Antimicrobial Susceptibility of Klebsiella pneumoniae from Community Settings in Taiwan, a Trend Analysis. Sci Rep 2016. [Google Scholar] [Crossref]
- D Maina, P Makau, A Nyerere, G Revathi. Antimicrobial resistance patterns in extended-spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae isolates in a private tertiary hospital, Kenya. Microbiol Discov 2013. [Google Scholar]
- VT Dougnon, K Sintondji, CH Koudokpon, M Houéto, AJ Agbankpé, P Assogba. Investigating Catheter-Related Infections in Southern Benin Hospitals: Identification, Susceptibility, and Resistance Genes of Involved Bacterial Strains. Microorganisms 2023. [Google Scholar] [Crossref]
- B Ashwak, WK Al-Musawy. Molecular Study and Antibiotic susceptibility patterns of some Extended Spectrum Beta-Lactamase Genes (ESBL) of Klebsiella pneumpniae in Urinary Tract Infections. J Phys Conf Ser 2020. [Google Scholar] [Crossref]
- T Engda, F Moges, A Gelaw, S Eshete, F Mekonnen. Prevalence and antimicrobial susceptibility patterns of extended spectrum beta-lactamase producing Entrobacteriaceae in the University of Gondar Referral Hospital environments, northwest Ethiopia. BMC Res Notes 2018. [Google Scholar] [Crossref]
- G Tayh, NA Al-Laham, I Fhoula, N Abedelateef, M El-Laham, AE Elottol. Frequency and Antibiotics Resistance of Extended-Spectrum Beta-Lactamase (ESBLs) Producing Escherichia coli and Klebsiella pneumoniae Isolated from Patients in Gaza Strip, Palestine. J Med Microbiol Infect Dis 2021. [Google Scholar]
- MB Jalil, MYN Al-Atbee. The prevalence of multiple drug resistance Escherichia coli and Klebsiella pneumoniae isolated from patients with urinary tract infections. J Clin Lab Anal 2022. [Google Scholar] [Crossref]
- S Subedi, M Chaudhary, B Shrestha. High MDR and ESBL Producing Escherichia coli and Klesbiella pneumoniae from Urine, Pus and Sputum Samples. Br J Med Med Res 2016. [Google Scholar]
- A Husna, MM Rahman, ATM Badruzzaman, MH Sikder, MR Islam, MT Rahman. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines 2023. [Google Scholar] [Crossref]
- S Verma, RK Kalyan, P Gupta, MD Khan, V Venkatesh. Molecular Characterization of Extended Spectrum β-Lactamase Producing Escherichia coli and Klebsiella pneumoniae Isolates and Their Antibiotic Resistance Profile in Health Care-Associated Urinary Tract Infections in North India. J Lab Physicians 2022. [Google Scholar]
- LI Thong, KT Lim, CC Yeo, SD Puthucheary, RM yasin. Genotypic Characterization of Extended-Spectrum β-lactamases Producing Klebsiella pneumoniae Strains Isolated in Malaysia. Int J Infect Dis 2008. [Google Scholar] [Crossref]
- G Odewale, MY Jibola-Shittu, O Ojurongbe, RA Olowe, OA Olowe. Genotypic Determination of Extended Spectrum β-Lactamases and Carbapenemase Production in Clinical Isolates of Klebsiella pneumoniae in Southwest Nigeria. Infect Dis Rep 2023. [Google Scholar]
- J Kopacz, N Mariano, R Colon-Urban, P Sychangco, W Wehbeh, S Segal-Maurer. Identification of extended-spectrum-β-lactamase-positive Klebsiella pneumoniae urinary tract isolates harboring KPC and CTX-M β-lactamases in non-hospitalized patients. Antimicrob Agents Chemother 2013. [Google Scholar]
- F Yao, Y Qian, S Chen, P Wang, Y Huang. Incidence of extended-spectrum beta-lactamases and characterization of integrons in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Shantou, China. Acta Biochim Biophys Sin (Shanghai) 2007. [Google Scholar]
- J Sun, F Zheng, F Wang, K Wu, Q Wang, Q Chen. Class 1 integrons in urinary isolates of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Southern China during the past five years. Microb Drug Resist 2013. [Google Scholar]
- IA Naqid, NR Hussein, AA Balatay, KA Saeed, HA Ahmed. The Antimicrobial Resistance Pattern of Klebsiella pneumoniae Isolated from the Clinical Specimens in Duhok City in Kurdistan Region of Iraq. J Kermanshah Univ Med Sci 2020. [Google Scholar] [Crossref]
- S Koirala, S Khadka, S Sapkota. Prevalence of CTX-M β-Lactamases Producing Multidrug Resistant Escherichia coli and Klebsiella pneumoniae among Patients Attending Bir Hospital, Nepal. Biomed Res Int 2021. [Google Scholar] [Crossref]
- A Taraghian, BN Esfahani, S Moghim, H Fazeli. Characterization of Hypervirulent Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Among Urinary Tract Infections: The First Report from Iran. Infect Drug Resist 2020. [Google Scholar] [Crossref]
- J Quan, H Dai, W Liao, D Zhao, Q Shi, L Zhang. Etiology and prevalence of ESBLs in adult community-onset urinary tract infections in East China: A prospective multicenter study. J Infect 2021. [Google Scholar]
- MH Dirar, NE Bilal, ME Ibrahim, ME Hamid. Prevalence of extended-spectrum β-lactamase (ESBL) and molecular detection of blaTEM, blaSHV and blaCTX-M genotypes among Enterobacteriaceae isolates from patients in Khartoum, Sudan. Pan Afr Med J 2020. [Google Scholar] [Crossref]
- AG Decano, K Pettigrew, W Sabiiti, DJ Sloan, S Neema, J Bazira. Pan-Resistome Characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae Strains Circulating in Uganda and Kenya. Antibiotics (Basel) 2021. [Google Scholar] [Crossref]
- KO Akinyemi, RO Abegunrin, BA Iwalokun, CO Fakorede, O Makarewicz, H Neubauer. The Emergence of Klebsiella pneumoniae with Reduced Susceptibility Against Third Generation Cephalosporins and Carbapenems in Lagos Hospitals. Antibiotics (Basel) 2021. [Google Scholar] [Crossref]
- S Vasaikar, L Obi, I Morobe, M Bisi-Johnson. Molecular Characteristics and Antibiotic Resistance Profiles of Klebsiella Isolates in Mthatha, Eastern Cape Province, South Africa. Int J Microbiol 2017. [Google Scholar] [Crossref]
- KSM Azab, MA Abdel-Rahman, HH El-Sheikh, E Azab, AA Gobouri, MMS Farag. Distribution of Extended-Spectrum β-Lactamase (ESBL)-Encoding Genes among Multidrug-Resistant Gram-Negative Pathogens Collected from Three Different Countries. Antibiotics (Basel) 2021. [Google Scholar] [Crossref]
- S H Elsafi. Occurrence and Characteristics of the Extended-spectrum Beta-lactamase Producing Enterobacterale in a Hospital Setting. Open Microbiol J 2020. [Google Scholar]
- Introduction
- Materials and Methods
- Study site
- Study design
- Study population
- Sample size determination
- Sampling criteria
- Specimen collection
- Urinalysis
- Urine culture
- Antimicrobial susceptibility test and ESBL detection
- DNA extraction
- Data and Statistical Analysis
- Results
- Demographic characteristics of study participants
- Dipstick positive and negative urine culture
- Prevalence of Klebsiella pneumoniae among UTI patients
- Antimicrobial susceptibility patterns
- Resistance genes of Klebsiella pneumoniae
- Discussion
- Socio-demographic characteristics of the participants
- Prevalence of Klebsiella pneumoniae among UTI patients
- Antimicrobial susceptibility patterns and ESBL producing K. pneumoniae
- Extended-spectrum β-lactamase resistance genes in Klebsiella pneumoniae
- Limitations
- Conclusion
- Ethical Consideration
- Conflicts of Interest
- Source of Funding
- Acknowledgments
